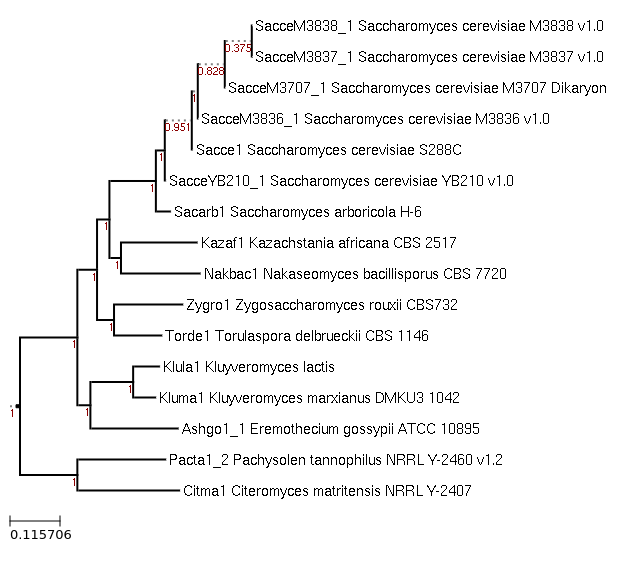
Maximum-Likelihood phylogeny generated by FastTree for
Kluyveromyces marxianus DMKU3 1042 and related species
The genome sequence and gene models of Kluyveromyces marxianus DMKU3 1042 were not determined by the Joint Genome Institute (JGI), but were downloaded from Ensembl Fungi on April 11, 2020. Please note that this copy of the genome is not maintained by Ensembl and is therefore not automatically updated. The JGI Annotation Pipeline was used to add additional functional annotation to the author's chromosomes and proteins.
Genome Reference(s)
Please cite the following publication(s) if you use the data from this genome in your research:
Lertwattanasakul N, Kosaka T, Hosoyama A, Suzuki Y, Rodrussamee N, Matsutani M, Murata M, Fujimoto N, Suprayogi, Tsuchikane K, Limtong S, Fujita N, Yamada M
Genetic basis of the highly efficient yeast Kluyveromyces marxianus: complete genome sequence and transcriptome analyses.
Biotechnol Biofuels. 2015;8():47. doi: 10.1186/s13068-015-0227-x
Lertwattanasakul N, Kosaka T, Hosoyama A, Suzuki Y, Rodrussamee N, Matsutani M, Murata M, Fujimoto N, Suprayogi, Tsuchikane K, Limtong S, Fujita N, Yamada M
Genetic basis of the highly efficient yeast Kluyveromyces marxianus: complete genome sequence and transcriptome analyses.
Biotechnol Biofuels. 2015;8():47. doi: 10.1186/s13068-015-0227-x
